We’re thrilled to announce the release of corrr 0.4.3. corrr is for exploring correlations in R. It focuses on creating and working with data frames of correlations (instead of matrices) that can be easily explored via corrr functions or by leveraging tools like those in the tidyverse.
You can install it from CRAN with:
install.packages("corrr")
This blog post will describe changes in the new version. You can see a full list of changes in the release notes.
library(corrr)
library(dplyr, warn.conflicts = FALSE)
Changes
This version of corrr has a few changes in the behavior of user-facing functions as well as the introduction of a new user-facing function.
There are also some internal changes that make package functions more robust. These changes don’t affect how you use the package but address some edge cases where previous versions were failing inappropriately.
New features of note are:
-
The first column of a
cor_dfobject is now named “term”. Previously it was named “rowname”. The name “term” is consistent with the output ofbroom::tidy(). This is a breaking change: code written to make use of the column name “rowname” will have to be amended. -
An
.orderargument has been added torplot()to allow users to choose the ordering of variables along the axes in the output plot. The default is that the output plots retain the variable ordering in the inputcor_dfobject. Setting.orderto “alphabet” orders the variables in alphabetical order in the plots. -
A new function,
colpair_map(), allows for column comparisons using the values returned by an arbitrary function.colpair_map()is discussed in detail below.
New column name in cor_df objects
We can create a cor_df object containing the pairwise correlations between a few numerical columns of the palmerpenguins::penguins data set to see that the first column is now named “term”:
library(palmerpenguins)
penguins_cor <- penguins %>%
select(bill_length_mm, bill_depth_mm, flipper_length_mm) %>%
correlate()
penguins_cor
## # A tibble: 3 x 4
## term bill_length_mm bill_depth_mm flipper_length_mm
## <chr> <dbl> <dbl> <dbl>
## 1 bill_length_mm NA -0.235 0.656
## 2 bill_depth_mm -0.235 NA -0.584
## 3 flipper_length_mm 0.656 -0.584 NA
Ordering variables in rplot() output
Previously, the default behavior of rplot() was that the variables were displayed in alphabetical order in the output. This was an artifact of using ggplot2 and inheriting its behavior. The new default is to retain the ordering of variables in the input data:
rplot(penguins_cor)
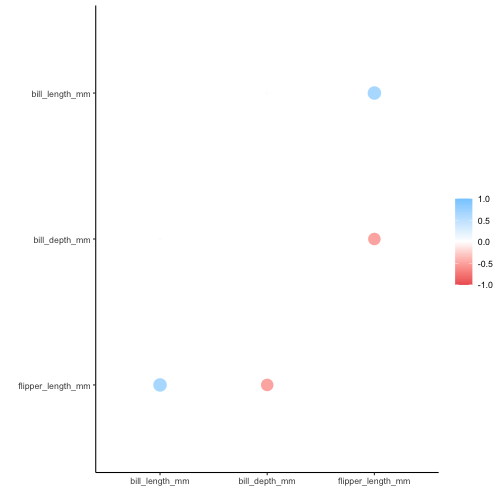
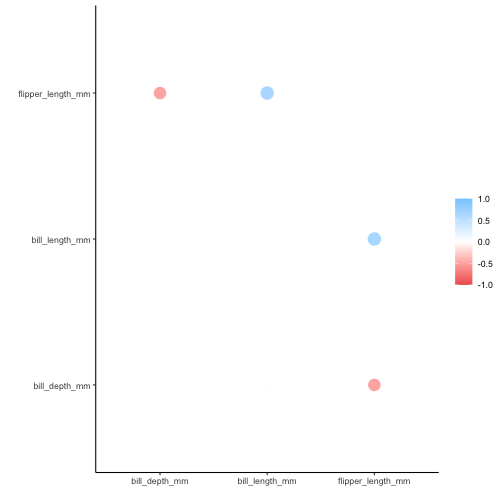
If alphabetical ordering is desired, set .order to “alphabet”:
rplot(penguins_cor, .order = "alphabet")
colpair_map()
Doing analysis with corrr has always been about correlations, usually starting with a call to correlate().
mini_mtcars <- mtcars %>% select(mpg, cyl, disp)
correlate(mini_mtcars)
##
## Correlation method: 'pearson'
## Missing treated using: 'pairwise.complete.obs'
## # A tibble: 3 x 4
## term mpg cyl disp
## <chr> <dbl> <dbl> <dbl>
## 1 mpg NA -0.852 -0.848
## 2 cyl -0.852 NA 0.902
## 3 disp -0.848 0.902 NA
The result is a data frame where each of the columns in the original data are compared on the basis of their correlation coefficients. But the correlation coefficient is just one possible statistic that can be used for comparing columns with one another. Correlations are also limited in their usefulness as they are only applicable to pairs of numeric columns.
This version of corrr introduces colpair_map(), which allows you to apply your own choice of function across the columns of your data. Just like with correlate(), colpair_map() takes a data frame as its first argument, while the second argument is for the function you wish to apply.
Let’s demonstrate using the mini_mtcars data frame we just created. Lets say we are interested in covariance values rather than correlations. These can be found by passing in cov() from the stats package:
cov_df <- colpair_map(mini_mtcars, stats::cov)
cov_df
## # A tibble: 3 x 4
## term mpg cyl disp
## <chr> <dbl> <dbl> <dbl>
## 1 mpg NA -9.17 -633.
## 2 cyl -9.17 NA 200.
## 3 disp -633. 200. NA
The resulting data frame behaves just like one returned by correlate(), except that it is populated with covariance values rather than correlations. This means we still have access to all corrr’s other tooling when working with it. We can still use shave() for example to remove duplication, which will set the upper triangle of values to NA.
cov_df %>%
shave()
## # A tibble: 3 x 4
## term mpg cyl disp
## <chr> <dbl> <dbl> <dbl>
## 1 mpg NA NA NA
## 2 cyl -9.17 NA NA
## 3 disp -633. 200. NA
Similarly, we can still use stretch() to get the resulting data frame into a longer format:
cov_df %>%
stretch()
## # A tibble: 9 x 3
## x y r
## <chr> <chr> <dbl>
## 1 mpg mpg NA
## 2 mpg cyl -9.17
## 3 mpg disp -633.
## 4 cyl mpg -9.17
## 5 cyl cyl NA
## 6 cyl disp 200.
## 7 disp mpg -633.
## 8 disp cyl 200.
## 9 disp disp NA
The first part of the name (“colpair_") comes from the fact that we are comparing pairs of columns. The second part of the name ("_map”) is designed to evoke the same ideas as in purrr’s family of map_* functions. These iterate over a set of elements and apply a function to each of them. In this case, colpair_map() is iterating over each possible pair of columns and applying a function to each pairing.
As such, any function passed to colpair_map() must accept a vector for both its first and second arguments. To illustrate, let’s say we wanted to run a series t-tests to see which of our variables are significantly related to one another. We can write a function to do so as follows:
calc_ttest_p_value <- function(vec_a, vec_b){
t.test(vec_a, vec_b)$p.value
}
The function returns the t-test’s p-value. The two arguments to the function are the two vectors being compared. Let’s first run the function on each pair of columns individually.
calc_ttest_p_value(mini_mtcars[, "mpg"], mini_mtcars[, "cyl"])
## [1] 9.507708e-15
calc_ttest_p_value(mini_mtcars[, "mpg"], mini_mtcars[, "disp"])
## [1] 7.978234e-11
calc_ttest_p_value(mini_mtcars[, "cyl"], mini_mtcars[, "disp"])
## [1] 1.774454e-11
As you can see, this is tedious and involves a lot of repeated code. But colpair_map() lets us do this for all column pairings at once and the output makes the results easy to read.
colpair_map(mini_mtcars, calc_ttest_p_value)
## # A tibble: 3 x 4
## term mpg cyl disp
## <chr> <dbl> <dbl> <dbl>
## 1 mpg NA 9.51e-15 7.98e-11
## 2 cyl 9.51e-15 NA 1.77e-11
## 3 disp 7.98e-11 1.77e-11 NA
Having the ability to use arbitrary functions like this opens up intriguing possibilities for analyzing data. One limitation of using only correlations is they will only work for continuous variables. With colpair_map(), we have a way of comparing categorical columns with one another. Let’s try this with a few categorical columns from dplyr’s dataset of Star Wars characters.
mini_star_wars <- starwars %>% select(hair_color, eye_color, skin_color)
head(mini_star_wars)
## # A tibble: 6 x 3
## hair_color eye_color skin_color
## <chr> <chr> <chr>
## 1 blond blue fair
## 2 <NA> yellow gold
## 3 <NA> red white, blue
## 4 none yellow white
## 5 brown brown light
## 6 brown, grey blue light
There are a few different ways of finding the strength of the relationship between two categorical variables. One useful measure is called Cramer’s V, which takes on values between 0 and 1 depending on how closely associated the variables are. The rcompanion package provides an implementation of Cramer’s V which we can make use of.
library(rcompanion)
colpair_map(mini_star_wars, cramerV)
## # A tibble: 3 x 4
## term hair_color eye_color skin_color
## <chr> <dbl> <dbl> <dbl>
## 1 hair_color NA 0.449 0.510
## 2 eye_color 0.449 NA 0.691
## 3 skin_color 0.510 0.691 NA
colpair_map() will allow you pass additional arguments to the called function via the dots (...). For example, the cramerV() function will allow you to specify the number of decimal places to round the results using digits. Let’s instead pass in this option via the dots:
colpair_map(mini_star_wars, cramerV, digits = 1)
## # A tibble: 3 x 4
## term hair_color eye_color skin_color
## <chr> <dbl> <dbl> <dbl>
## 1 hair_color NA 0.4 0.5
## 2 eye_color 0.4 NA 0.7
## 3 skin_color 0.5 0.7 NA
We are excited to see the different ways colpair_map() gets used by the R community. We are hopeful that it will open up new and exciting ways of conducting data analysis.
Acknowledgements
We’d like to thank everyone who contributed to the package or filed an issue since the last release: @Aariq, @antoine-sachet, @bjornerstedt, @jameslairdsmith, @jamesMo84, @juliangkr, @juliasilge, @mattwarkentin, @mwilson19, @norhther, @thisisdaryn, and @topepo.
