readr 1.3.1 is now available on CRAN! Learn more about readr at https://readr.tidyverse.org. Detailed notes are always in the change log.
The readr package makes it easy to get rectangular data out of comma separated (csv), tab separated (tsv) or fixed width files (fwf) and into R. It is designed to flexibly parse many types of data found in the wild, while still cleanly failing when data unexpectedly changes. If you are new to readr, the best place to start is the data import chapter in R for data science.
The easiest way to install the latest version from CRAN is to install the whole tidyverse.
install.packages("tidyverse")
Alternatively, install just readr from CRAN:
install.packages("readr")
readr is part of the core tidyverse, so load it with:
library(tidyverse)
#> ── Attaching packages ────────────────────────────────── tidyverse 1.2.1 ──
#> ✔ ggplot2 3.1.0 ✔ purrr 0.2.5
#> ✔ tibble 1.4.2 ✔ dplyr 0.7.8
#> ✔ tidyr 0.8.2 ✔ stringr 1.3.1
#> ✔ readr 1.3.1 ✔ forcats 0.3.0
#> ── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
There have been three releases of readr recently, this post will highlight the most significant changes across all three releases.
Breaking Changes
Blank line skipping
The logic for blank line skipping has been overhauled, which should make it easier to predict and more robust.
Now both commented lines and completely blank lines are skipped by default.
file <-
"# A file with comments
first_name,last_name
Mary,Sue
John,Doe
"
read_csv(file, comment = "#")
#> # A tibble: 2 x 2
#> first_name last_name
#> <chr> <chr>
#> 1 Mary Sue
#> 2 John Doe
Some useful properties about skip.
- It is a lower bound on the number of lines that will actually be skipped. e.g. both of the following examples result in identical output.
read_csv(file, skip = 1)
#> # A tibble: 2 x 2
#> first_name last_name
#> <chr> <chr>
#> 1 Mary Sue
#> 2 John Doe
read_csv(file, skip = 2)
#> # A tibble: 2 x 2
#> first_name last_name
#> <chr> <chr>
#> 1 Mary Sue
#> 2 John Doe
skipis enforced without looking at the line contents, so both commented and blank lines are counted.
So if you wanted to skip all the way including the first_name,last_name line
you would need to use skip = 3. N.B. the results are identical with and
without comment = "#".
read_csv(file, skip = 3, col_names = FALSE)
#> # A tibble: 2 x 2
#> X1 X2
#> <chr> <chr>
#> 1 Mary Sue
#> 2 John Doe
Integer column guessing
readr functions no longer guess columns are of type integer, instead these
columns are guessed as numeric. Because R uses 32 bit integers and 64 bit
doubles all integers can be stored in doubles, guaranteeing no loss of
information. This change was made to remove errors when numeric columns were
incorrectly guessed as integers. If you know a certain column is an integer and
would like to read it as such you can do so by specifying the column type
explicitly with the col_types argument.
file <-
"a,b
1,3
2,4
"
read_csv(file, col_type = c(b = "i"))
#> # A tibble: 2 x 2
#> a b
#> <dbl> <int>
#> 1 1 3
#> 2 2 4
tibble subclass
readr now returns results with a spec_tbl_df subclass. This differs from a
regular tibble only in that the spec attribute (which holds the column
specification) is lost as soon as the object is subset and a normal tbl_df
object is returned.
Historically tbl_df's lost their attributes once they were subset. However
recent versions of tibble retain the attributes when subsetting, so the
spec_tbl_df subclass is needed to retain the previous behavior.
This should only break compatibility if you are explicitly checking the class
of the returned object. A way to get backwards compatible behavior is to
call subset with no arguments on your object, e.g. x[].
data <- read_csv(file)
class(data)
#> [1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"
class(data[])
#> [1] "tbl_df" "tbl" "data.frame"
New features
Colored specifications
The most user visible change is coloration of the column specifications. The column types are now colored based on 4 broad classes
- Red - Character data (characters, factors)
- Green - Numeric data (doubles, integers)
- Yellow - Logical data (logicals)
- Blue - Temporal data (dates, times, datetimes)
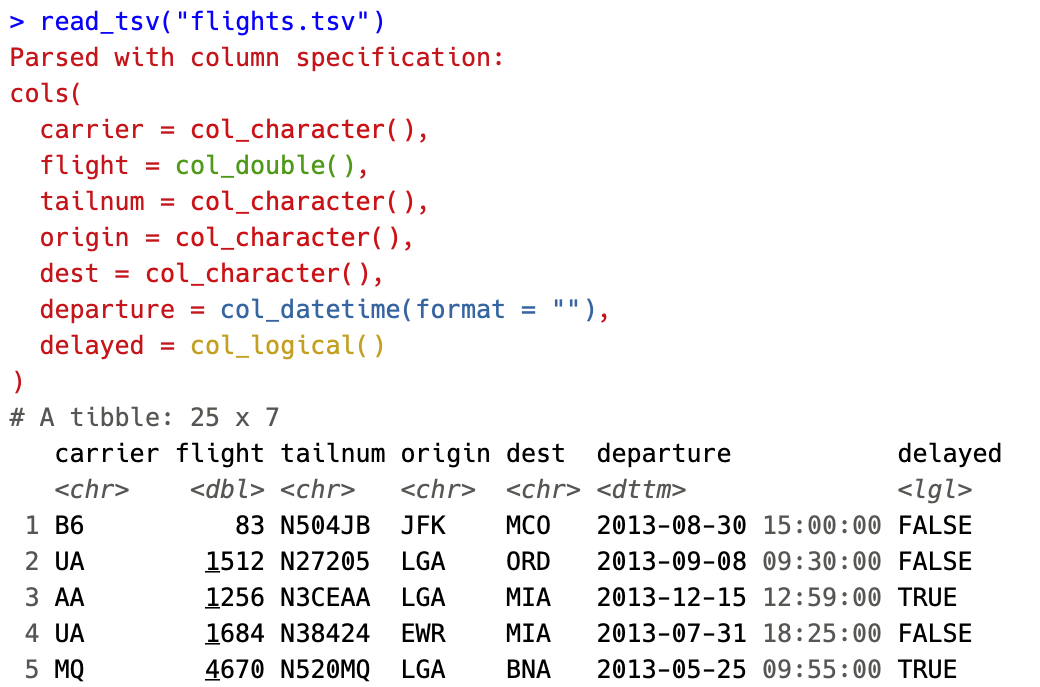
By coloring the specification, we hope to make it easier to spot when a column differs from the rest or when guessing leads to import with an unexpected type.
The coloring can be disabled by setting options(crayon.enabled = FALSE).
Melt functions
There is now a family of melt_*() functions in readr. These functions store
data in ‘long’ or ‘melted’ form, where each row corresponds to a single
value in the dataset. This form is useful when your data is ragged and not
rectangular.
file <-
"a,b,c
1,2
w,x,y,z"
readr::melt_csv(file)
#> # A tibble: 9 x 4
#> row col data_type value
#> <dbl> <dbl> <chr> <chr>
#> 1 1 1 character a
#> 2 1 2 character b
#> 3 1 3 character c
#> 4 2 1 integer 1
#> 5 2 2 integer 2
#> 6 3 1 character w
#> 7 3 2 character x
#> 8 3 3 character y
#> 9 3 4 character z
Thanks to
Duncan Garmonsway (@nacnudus) for great work on the idea and
implementation of the melt_*() functions!
Connection improvements
In previous versions of readr the connections were read into an in-memory raw vector, then passed to the readr functions. This made reading connections for small to medium datasets fast, but also meant that the dataset had to fit into memory at least twice (once for the raw data, once for the parsed data). It also meant that reading could not begin until the full vector was read through the connection.
We now write the connection to a temporary file (in the R temporary directory), than parse that temporary file. This means connections may take a little longer to be read, but also means they will no longer need to fit into memory. It also allows the use of the chunked readers to process the data in parts.
Future improvements to readr could allow it to parse data from connections in a streaming fashion, which would avoid many of the drawbacks of either approach.
Acknowledgements
Thank you to the 188 contributors who made this release possible by opening issues or submitting pull requests: @aaronschiff, @abelew, @Abhijitsj, @ADraginda, @adrtod, @ajdamico, @alessandro-gentilini, @arendsee, @batpigandme, @bbrewington, @benmarwick, @benwhalley, @benzipperer, @BerwinTurlach, @bheavner, @billdenney, @biostu24, @blablablerg, @blakeboswell, @brainfood, @brianstamper, @brisk022, @Brunohnp, @bschneidr, @cameron-faerber, @Carenser, @cboettig, @cderv, @ChiWPak, @christellacaze, @cimentadaj, @conchoecia, @cuttlefish44, @dan-reznik, @danielsjf, @dapperjapper, @DarioS, @DataStrategist, @DDWetzel, @DevGri, @dfjenkins3, @dhbrand, @dhimmel, @digital-dharma, @dioh, @djbirke, @dkulp2, @dongzhuoer, @dpprdan, @Dulani, @eddelbuettel, @ElToro1966, @EmilHvitfeldt, @Enchufa2, @epetrovski, @eric-pedersen, @EricHe98, @EruIluvatar, @evelynmitchell, @EvgenyPetrovsky, @gandalf-randolph, @GegznaV, @gergness, @ghost, @gksmyth, @gregrs-uk, @gwarnes-mdsol, @ha0ye, @hadley, @hbeale, @Henri-Lo, @hiltonmbr, @hmorzaria, @holstius, @hstojic, @huftis, @ilyasustun, @IndrajeetPatil, @isomorphisms, @isteves, @j-bartholome, @J535D165, @jaearick, @jasonserviss, @javierluraschi, @jdeboer, @jennybc, @jhpoelen, @jiho, @jimhester, @jimmo, @johncassil, @jomuller, @jonovik, @josiekre, @jpiggod, @jschelbert, @jstaf, @jtelleria, @jtelleriar, @jtr13, @juyeongkim, @jzadra, @kadekillary, @kendonB, @kerry-ja, @kevinushey, @kforner, @kk22boy, @KKulma, @kos59125, @kwstat, @laresbernardo, @lbartnik, @ldecicco-USGS, @Leo-Lee15, @lsusatyo-gcmlp, @martinjhnhadley, @martj42, @matthewarbo, @MatthieuStigler, @mawds, @md0u80c9, @mgirlich, @mikmart, @mirkhosro, @mohkar123, @momeara, @msberends, @msmall318, @Naboum, @nacnudus, @namelessjon, @nan1949, @nbenn, @neveldo, @nevrome, @NicolasImberty, @normandev, @olgamie, @osorensen, @pachevalier, @pakuipers, @pgensler, @PMassicotte, @prebours, @prosoitos, @QuLogic, @rafazaya, @ramiromagno, @randomgambit, @raw937, @rdisalv2, @rensa, @richierocks, @RickPack, @rmcd1024, @RomeroBarata, @roshnamohan, @ryanatanner, @s-fleck, @saghirb, @sambrady3, @seandavi, @seanpor, @sellisd, @sibojan, @SimonGoring, @sindile, @statsccpr, @stelsemeyer, @StevenMMortimer, @stianlagstad, @tarmenic, @tdsmith, @thanosgatos, @tigertoes, @tomsing1, @TPreijers, @tres-pitt, @tungmilan, @wgrundlingh, @wibeasley, @willemvdb42, @xhudik, @yutannihilation, @Zack-83, and @zeehio
